Evoluce kvazidruhu za přítomnosti mutací
![]()
![]()
Konstanty modelu:
![]()
| (1) |
Zdatnost:
![]()
| (2) |
Archivace střední hodnoty a rozptylu:  Vykreslit zdatnost
Vykreslit zdatnost
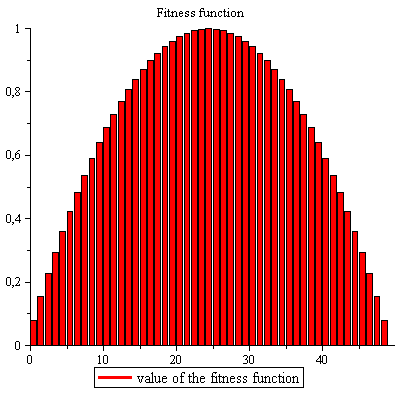 Populace v procentech, celkově 100%:
Populace v procentech, celkově 100%:
![]()
![]()
![]()
![]()

(3)
![]()
(4)
 Procedura na vykreslení sloupc
Procedura na vykreslení sloupc
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
 Procedura pro vývoj v jednom č
Procedura pro vývoj v jednom č
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
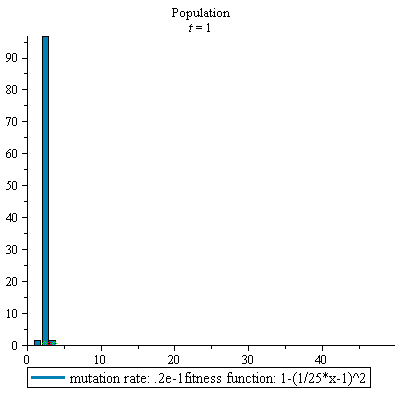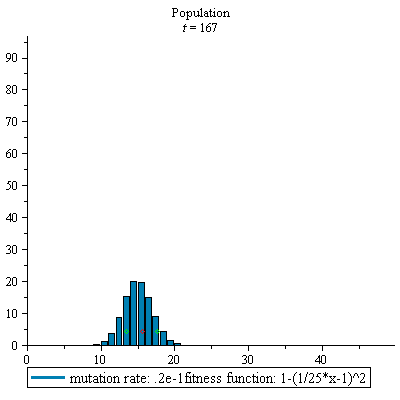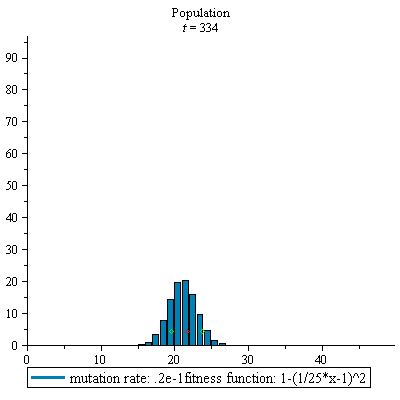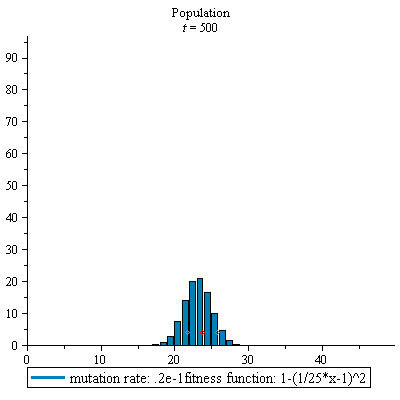
 display(
display(
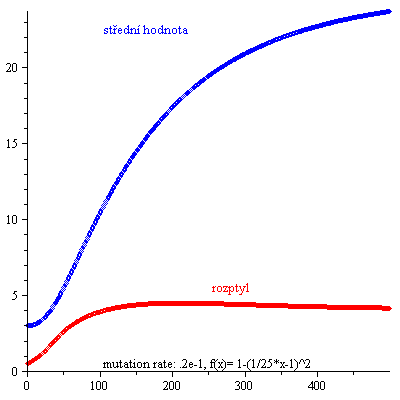
 Kontrolní součet
Kontrolní součet
![]()
![]()